#
Dr. M. Baron, Statistical Machine Learning class, STAT-427/627
# CROSS-VALIDATION
1. Validation Set Approach
> library(ISLR2)
> attach(Auto); n = length(mpg);
> n
[1] 392
> Z = sample( n, 250 ) # Random subsample of size 250 (does not
have to be n/2)
# Works similarly to generating a Bernoulli
variable
> reg.fit = lm( mpg ~ weight + horsepower + acceleration, subset=Z ) # Fit using training data
> mpg_predicted =
predict( reg.fit, newdata=Auto[-Z
, ] )
#
Use this model to predict the testing data [-Z]
> plot(mpg[-Z],mpg_predicted) # We can see a nonlinear component
> abline(0,1) # Compare with the line y=x.
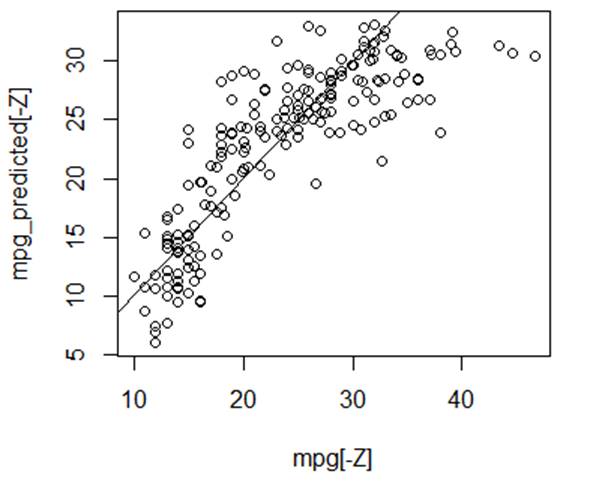
> mean( (mpg[-Z] - mpg_predicted)^2 ) # Estimate
the mean-squared error MSE
[1] 18.58128
2. Jackknife (Leave-One-Out Cross-Validation,
LOOCV)
“Manually”:
> Yhat = numeric(n)
> for (i in 1:n){
+ reg = lm( mpg ~ weight + horsepower + acceleration,
data=Auto[-i,] )
+ Yhat[i]
= predict( reg, newdata=Auto[i,] )
+ }
> plot(mpg,Yhat)
> mean((mpg-Yhat)^2)
[1] 18.25595
Using package “boot”:
> install.packages(“boot”)
> library(boot)
> glm.fit = glm(mpg ~ weight + horsepower + acceleration) #
Default “family” is Normal, so this GLM model
> cv.error = cv.glm(
Auto, glm.fit )
# is the same as LM, standard linear
regression.
> names(cv.error)
# This cross-validation tool has several
outputs
[1] "call" "K" "delta" "seed" # We are interested in “delta”
> error$delta
[1] 18.25595 18.25542
# Delta consists of 2 numbers – estimated prediction error and its
version adjusted for the lost sample size due to cross-validation.
# Example – what power of “horsepower” is optimal for this
prediction?
> cv.error =
rep(0,10) # Initiate a vector of estimated errors
> for (p in 1:10){ # Fit polynomial regression models
+ glm.fit = glm( mpg ~ weight + poly(horsepower,p)
+ acceleration ) # with power p=1..10
+ cv.error[p] = cv.glm( Auto, glm.fit
)$delta[1] } # Save prediction errors
> cv.error # Look at the results
[1]
18.25595 15.90163 15.72995 15.86879 15.74517 15.74989 15.70073 15.79314
15.95933 16.38301
# Although p=7 yields the lowest estimated prediction error, after
p=2, the improvement is very little.
> plot(cv.error)
> lines(cv.error)
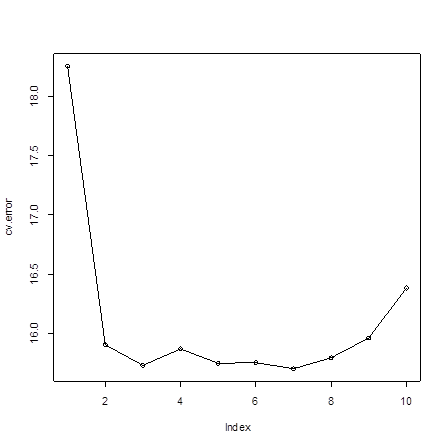
3. K-Fold Cross-Validation
# We can specify K within cv.glm (Omitted K
is K=1 by default, which is LOOCV).
> cv.error =
rep(0,10)
> for (p in 1:10){ glm.fit
= glm( mpg ~ weight + poly(horsepower,p)
+ acceleration )
+ cv.error[p] = cv.glm( Auto, glm.fit, K=30 )$delta[1] }
> cv.error
[1]
18.22699 15.87030 15.73565 15.85911 15.80686 15.71585 16.09625 15.67797
15.82097 16.48196
> which.min(cv.error)
[1] 6
4. Cross-validation in classification problems.
Loss function.
# Command cv.glm, as we used it above,
calculates MSE, the mean squared error, and calls them “delta”.
# In classification problems, MSE can be used to measure the distance
between the response variable Y
# and the predicted probability p. For this, Y has
to be 0 or 1, and p should be the probability of Y=1.
# However, the correct classification rate and the error
classification rate are more standard measures
# of classification accuracy. We can force cv.glm
to return these measures by introducing a suitable loss
# function. For example, we’ll be predicting whether a student is
depressed or not.
# We define a
loss function L(Y,p), a function of true
response Y and predicted probability p. The loss = 1 if
# the predicted response is different from
the actual response and 0 otherwise. Suppose the threshold is 0.5
> loss = function(Y,p){
return( mean( (Y==1 & p < 0.5) | (Y==0 & p >= 0.5) ) ) }
> loss(1,0.3)
[1] 1
> loss(c(1,1),c(0.3,0.7))
[1] 0.5
# Now we attach the Depression data, skip missing values, fit
logistic regression model, and estimate
# the error classification rate by LOOCV.
> Depr =
read.csv(url("http://fs2.american.edu/~baron/627/R/depression_data.csv"))
> D = na.omit(Depr)
> attach(D)
> library(boot)
> lreg = glm(Diagnosis ~ Gender + Guardian_status
+ Cohesion_score, family="binomial")
> cv = cv.glm( D, lreg, loss )
> cv$delta[1]
[1]
0.1615721
# Is Guardian_status significant?
Let’s compare the error rates with and without variable “Guardian_status”.
> lreg = glm(Diagnosis ~ Gender + Cohesion_score,
family="binomial")
> cv.glm( D, lreg, loss )$delta[1]
[1] 0.1572052
# The error classification rate is lower without the “Guardian_status”.